The Cleavage Under Targets & Release Using Nuclease (CUT&RUN) assay is a faster, more cost-effective alternative to ChIP-qPCR and ChIP-seq that requires fewer cells to investigate protein-DNA interactions. Cell Signaling Technology® (CST®) has made its CUT&RUN Kit even better by performing additional validation and optimization to improve the protocol, resulting in even more options for how you explore protein-DNA interactions.
CUT&RUN is an in vivo method that delivers results in 1-2 days, lowers sequencing costs, and is compatible with a variety of antibodies and targets. The CUT&RUN Assay Kit now offers these additional benefits to increase assay flexibility:
| Improved protocols | Optimized protocols enhance the enrichment of low abundance and/or weak binding transcription factors and cofactors |
| Even lower sample requirement | 5-10K cells for Histones 10-20K cells for Transcription Factors and Cofactors |
| Now compatible with fixed cells | Light cell fixation keeps cells intact, preserves cell signaling pathways, and enhances the enrichment for accessory components of huge complexes |
| Use fixed or fresh tissue samples with confidence | Validated and optimized tissue protocols give you data confidence when using ~20x less sample compared to ChIP |
| Study protein-DNA interactions in primary cells | CUT&RUN significantly lowers the cell number requirements, making it suitable to use with primary cells |
Plus, don’t forget that the CUT&RUN Assay Kit from CST provides:
| Fast time to results | 1-2 days from cell to DNA |
| Lower sequencing depth = lower sequencing costs | Only requires 3-5 million high-quality reads per sample due to the inherently low assay background |
| In Vivo Method | Assays are performed using native chromatin, eliminating cross-linking artifacts |
| Antibody versatility | Compatible with rabbit and mouse antibodies |
| Target versatility | Generate sequencing and/or qPCR data for histones, histone modifications, transcription factors, and cofactors. |
| Straightforward quantification | Spike in control DNA to simplify data quantification and normalization. |
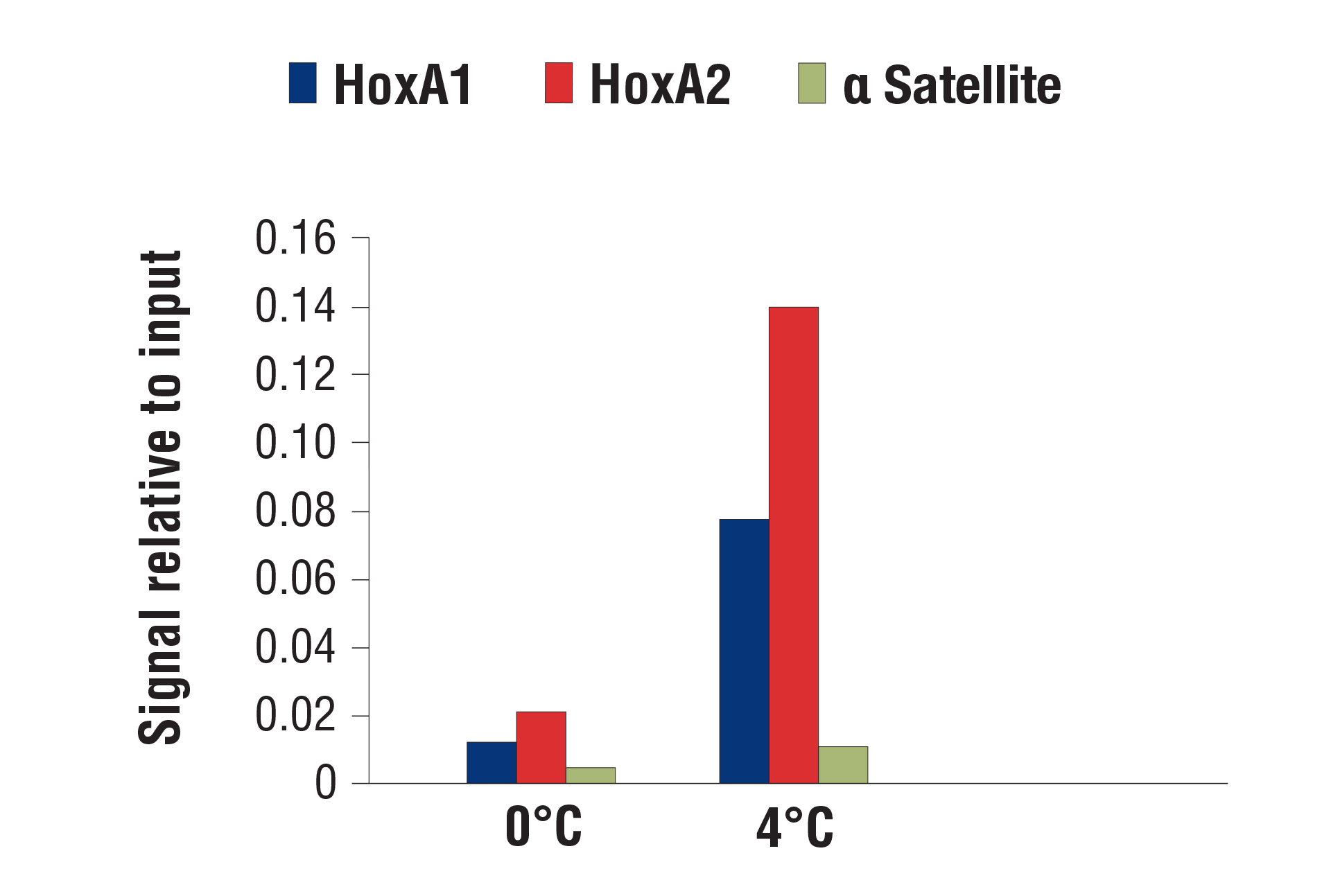
Continued development on the CUT&RUN protocol has further optimized the protocol to enhance your data and validated the CUT&RUN Assay Kit for use with a wider variety of sample types. For example, we now recommend performing digestions at 4°C instead of 0°C to facilitate the recovery of targeted chromatin fragments without significantly increasing background. This improves qPCR and NG-seq analysis, especially for transcription factors and cofactors.
Panel A
Panel B
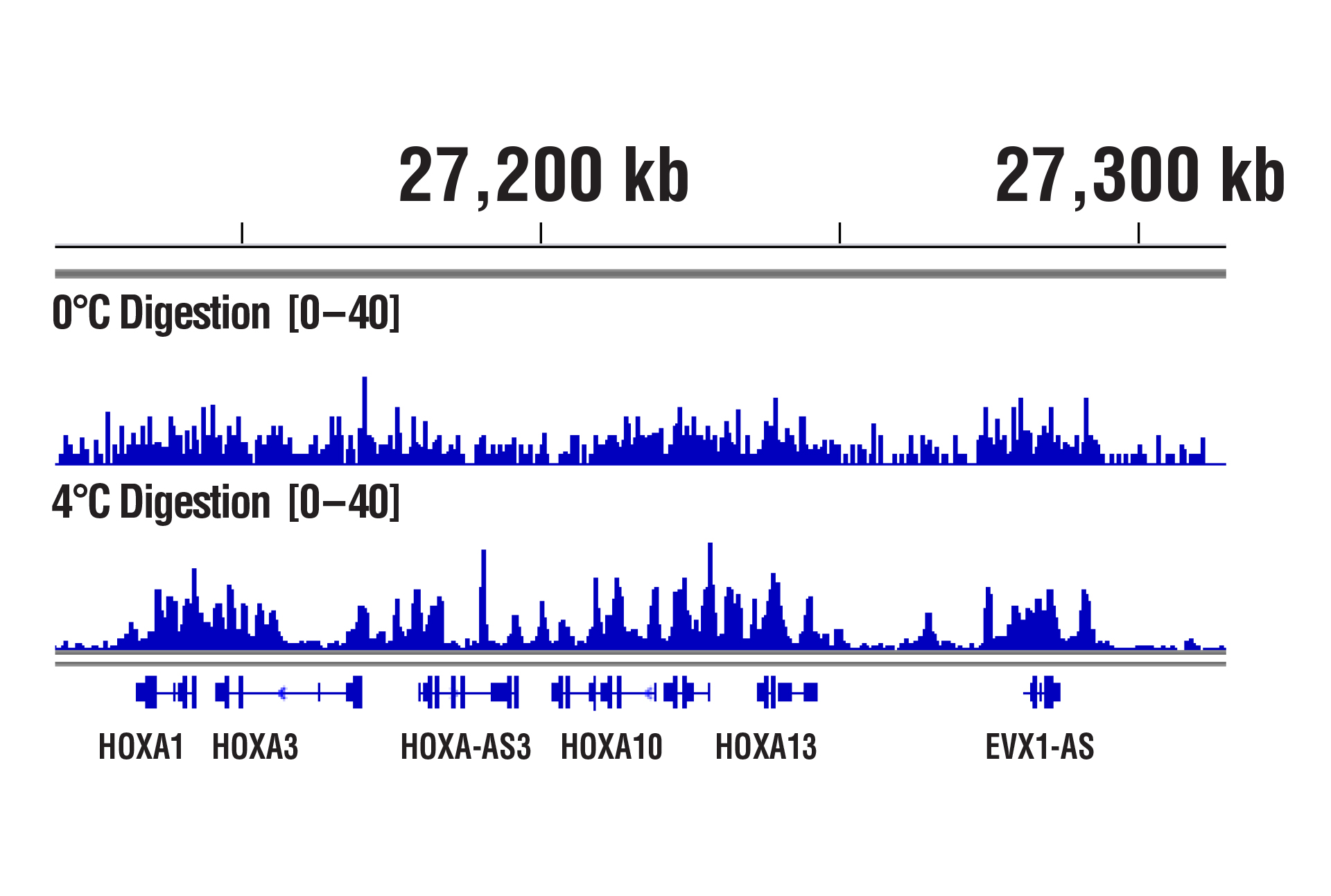
CUT&RUN was performed with NCCIT cells and SUZ12 (D39F6) XP® Rabbit mAb #3737, using CUT&RUN Assay Kit #86652. DNA was digested at either 0°C or 4°C as indicated. In panel A, the enriched DNA was quantified by real-time PCR using SimpleChIP® Human HoxA1 Intron 1 Primers #7707, SimpleChIP® Human HoxA2 Promoter Primers #5517, and SimpleChIP® Human α Satellite Repeat Primers #4486. The amount of immunoprecipitated DNA in each sample is represented as signal relative to the total amount of input chromatin, which is equivalent to one. In panel B, DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. The figure shows binding across HoxA genes.
CUT&RUN already required significantly fewer cells to generate reliable data compared to ChIP-qPCR and ChIP-seq, and CST has further increased your time savings by optimizing protocols to use even less sample.
Unable to collect 100K cells, but still want to perform CUT&RUN? Not a problem as you can generate the same great data with just 5-20K cells.
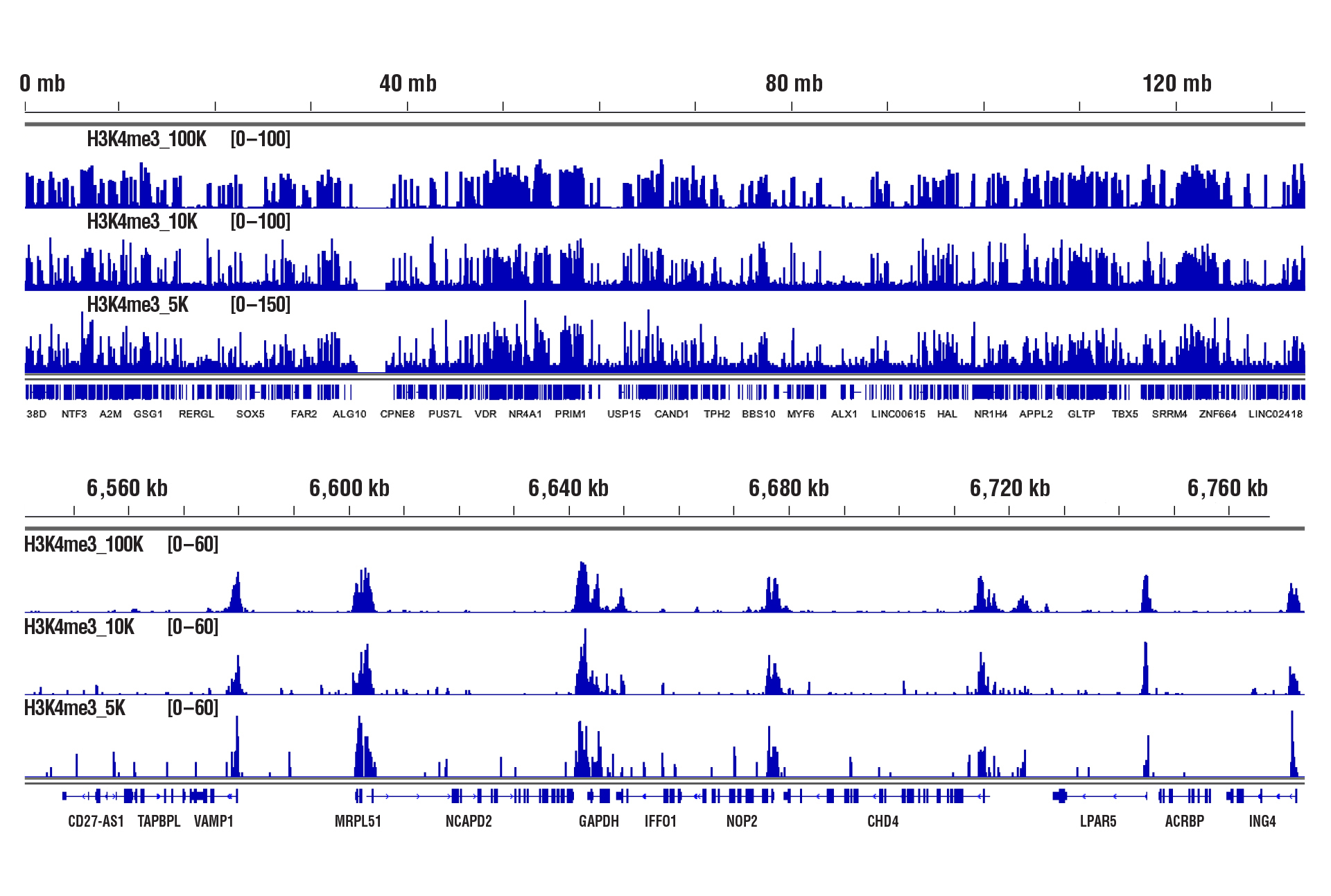
Similar binding patterns were observed when 100,000, 10,000, or 5,000 cells were used on the CUT&RUN Assay Kit #86652 when studying histones.
Panel A
Panel B
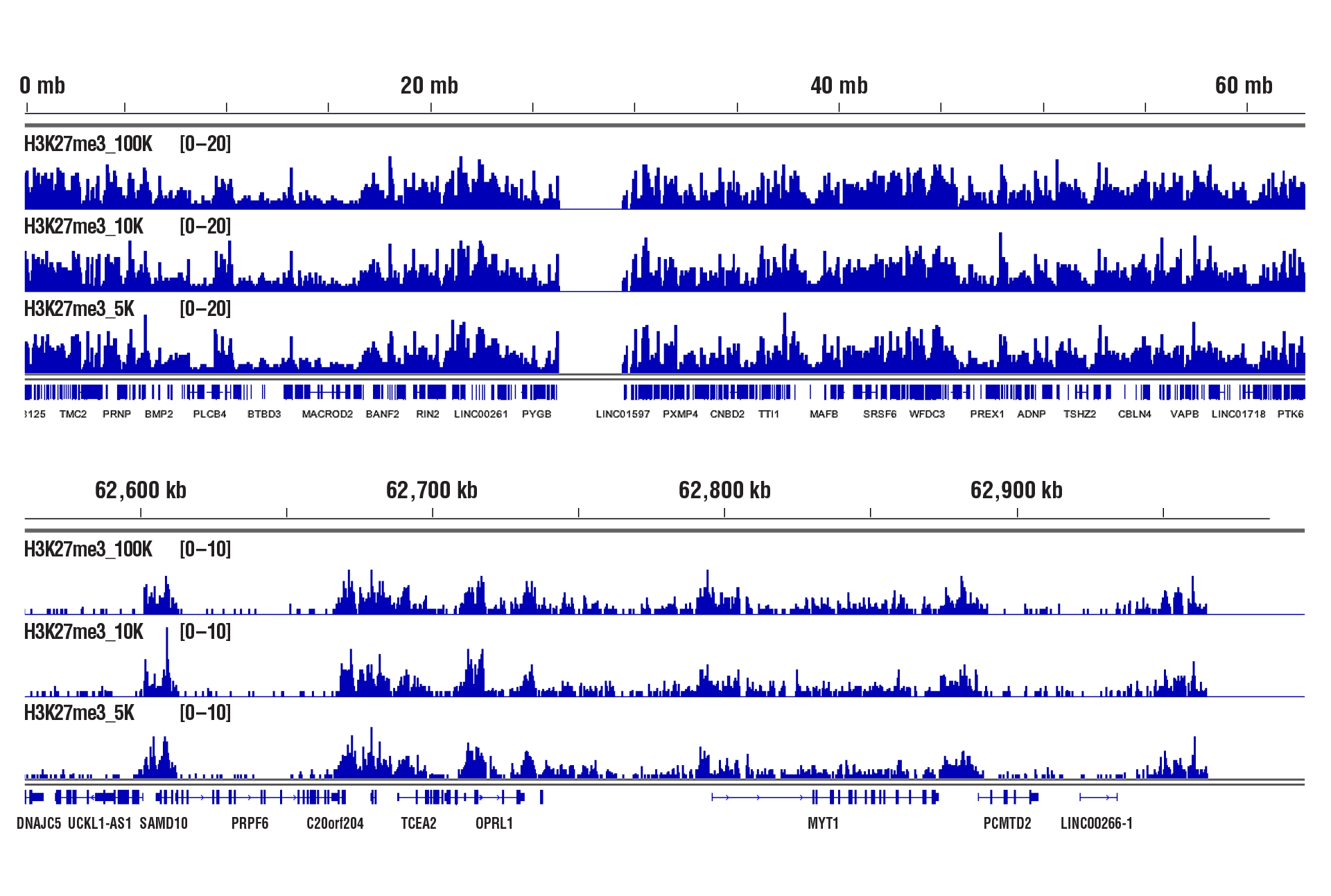
CUT&RUN was performed with decreasing numbers of cells using CUT&RUN Assay Kit #86652. In panel A, 100,000, 10,000, or 5,000 HCT 116 cells and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. The figures show binding across chromosome 12 (upper), including GAPDH (lower), a known target gene of H3K4me3. In panel B, CUT&RUN was performed with 100,000, 10,000, or 5,000 HeLa cells (as indicated) and Tri-Methyl-Histone H3 (Lys27) (C36B11) Rabbit mAb #9733. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. The figures show binding across chromosome 20 (upper), including MYT1 (lower), a known target gene of H3K27me3.
Similar binding patterns were observed when 100,000, 20,000, or 10,000 cells were used on the CUT&RUN Assay Kit #86652 when studying transcription factors.
Panel A
Panel B
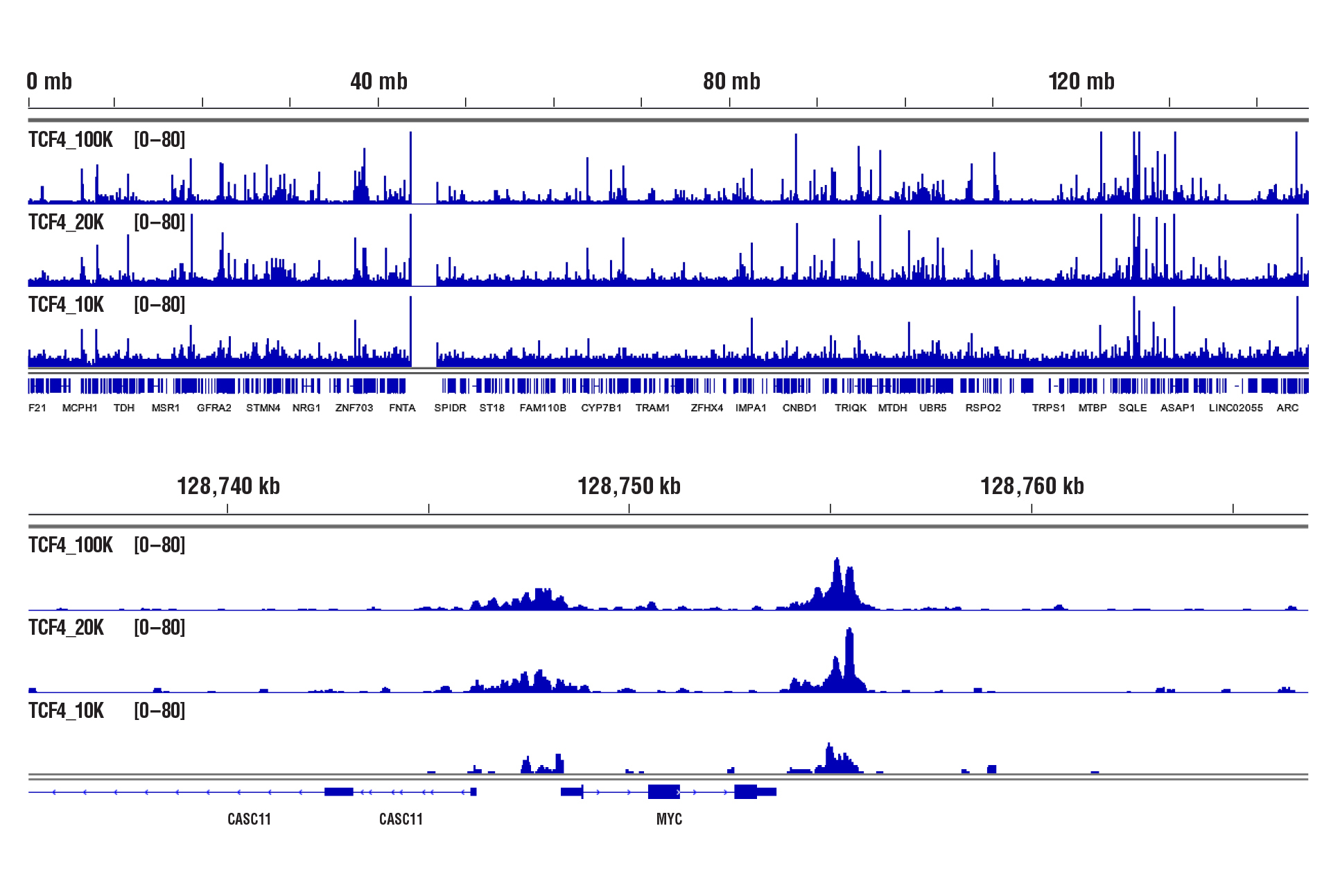
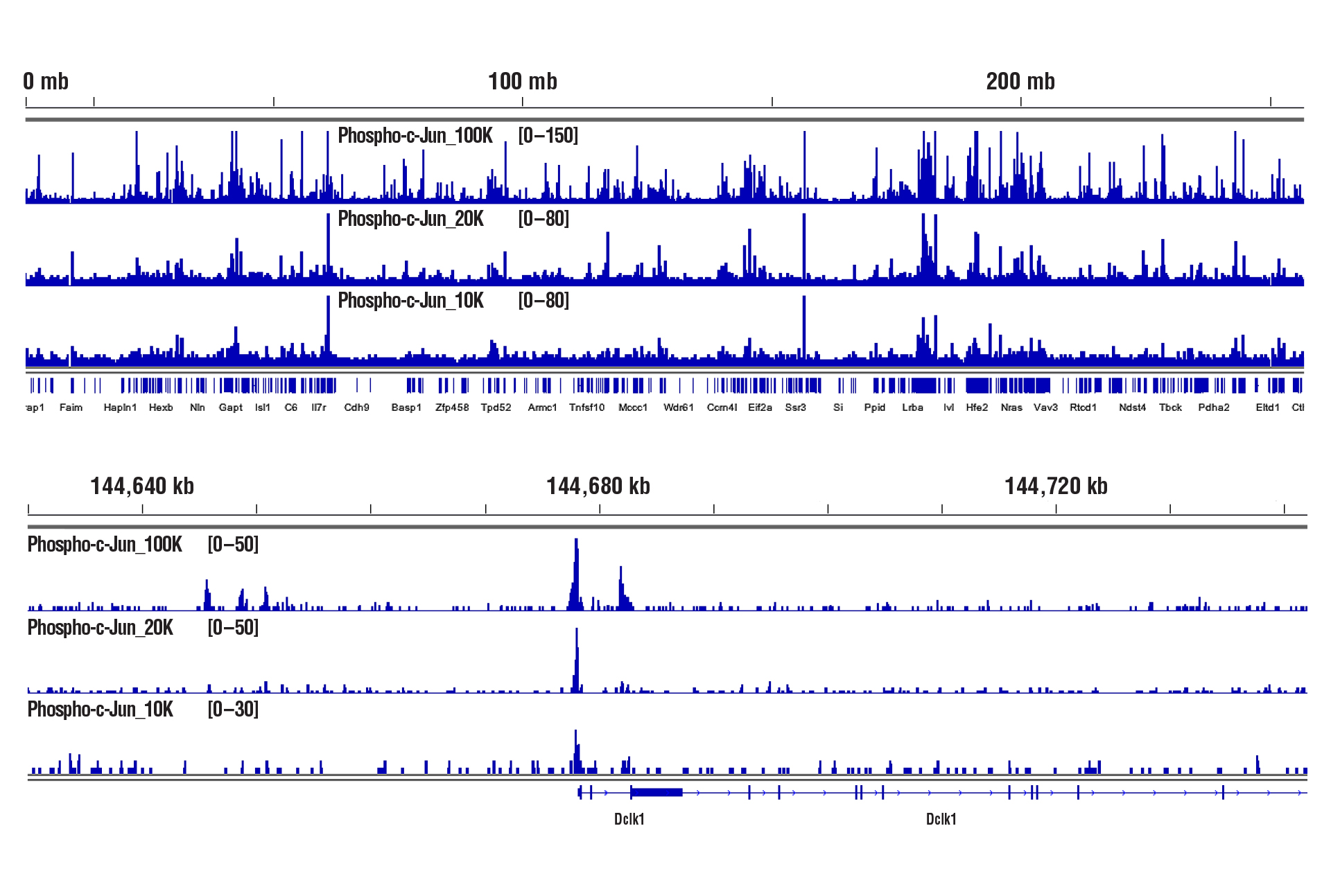
CUT&RUN was performed with decreasing numbers of cells using CUT&RUN Assay Kit #86652. In panel A, CUT&RUN was performed with 100,000, 20,000, or 10,000 PC-12 starved overnight and treated with Human β-Nerve Growth Factor (hβ-NGF) #5221 (50 ng/mL) for 2 hrs and Phospho-c-Jun (Ser73) (D47G9) XP® Rabbit mAb #3270. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. The figure shows binding across chromosome 2 (upper), including Dclk1 (lower), a known target gene of Phospho-c-Jun. In panel B, CUT&RUN was performed with 100,000, 20,000, or 10,000 HCT 116 cells and TCF4/TCF7L2 (C48H11) Rabbit mAb #2569. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. The figures show binding across chromosome 8 (upper), including MYC (lower), a known target gene of TCF4.
Similar binding patterns were observed when 100,000, 20,000, or 10,000 cells were used on the CUT&RUN Assay Kit #86652 when studying cofactors.
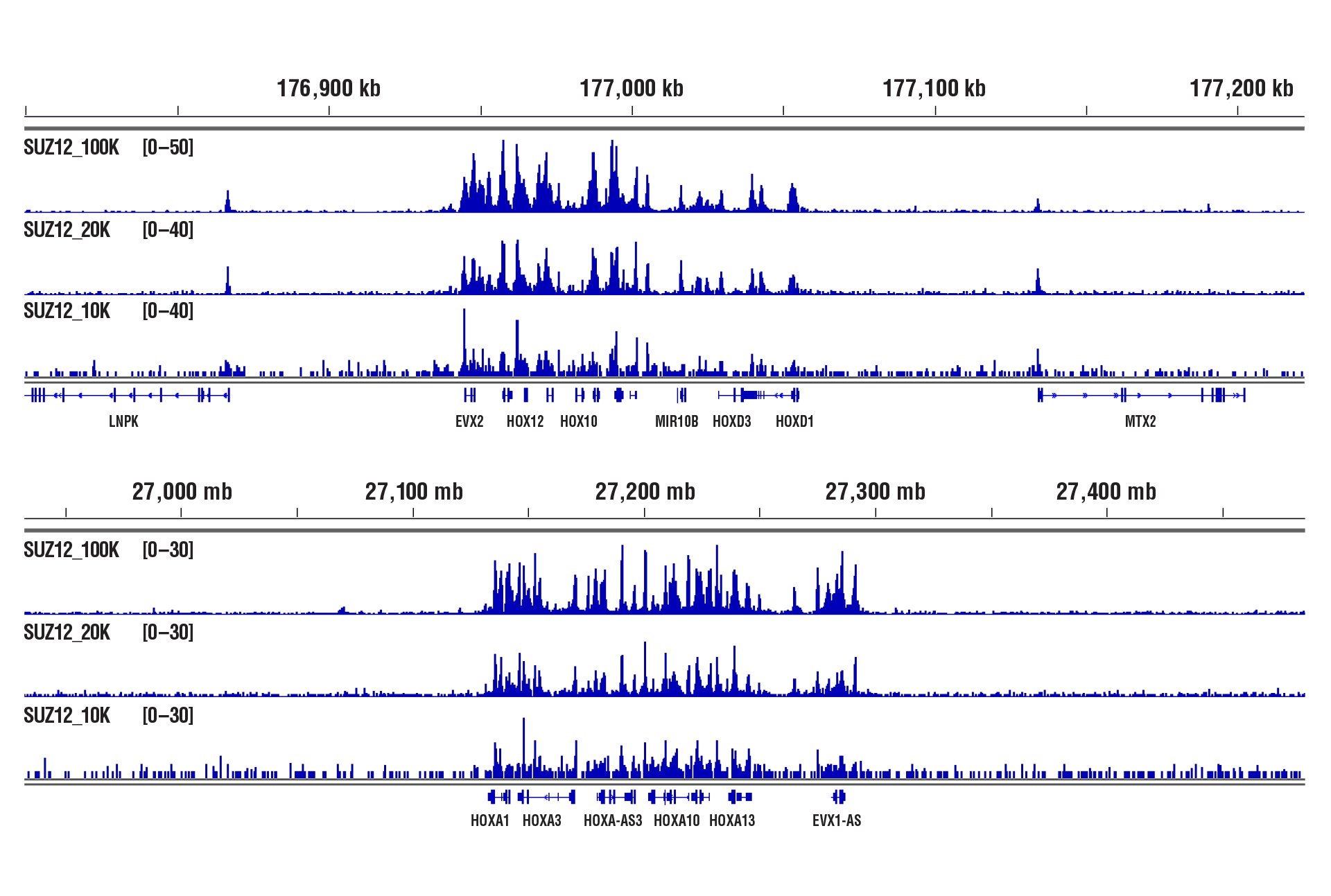
CUT&RUN was performed with 100,000, 20,000, or 10,000 NCCIT cells and SUZ12 (D39F6) XP® Rabbit mAb #3737, using CUT&RUN Assay Kit #86652. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. The figures show binding across HoxD (upper) and HoxA (lower), a known target gene of SUZ12.
The high sample input required to perform ChIP experiments is extremely restrictive when it comes to profiling chromatin in limited sample types like tissue. The sample savings CUT&RUN delivers for cultured cells extends to tissue samples. Depending on the tissue, ChIP can require anywhere from 25 to 50 mg of tissue while our CUT&RUN kit now only requires approximately 1 to 2.5 mg depending on the tissue type.
Validated protocols give you confidence in your data even when using ~20x less sample to explore protein-DNA interactions using the CST CUT&RUN Assay Kit #86652.
Only 1 mg of heart tissue was required to analyze protein-DNA interactions for H3K4me3 and transcription factor MITF using our CUT&RUN Kit.
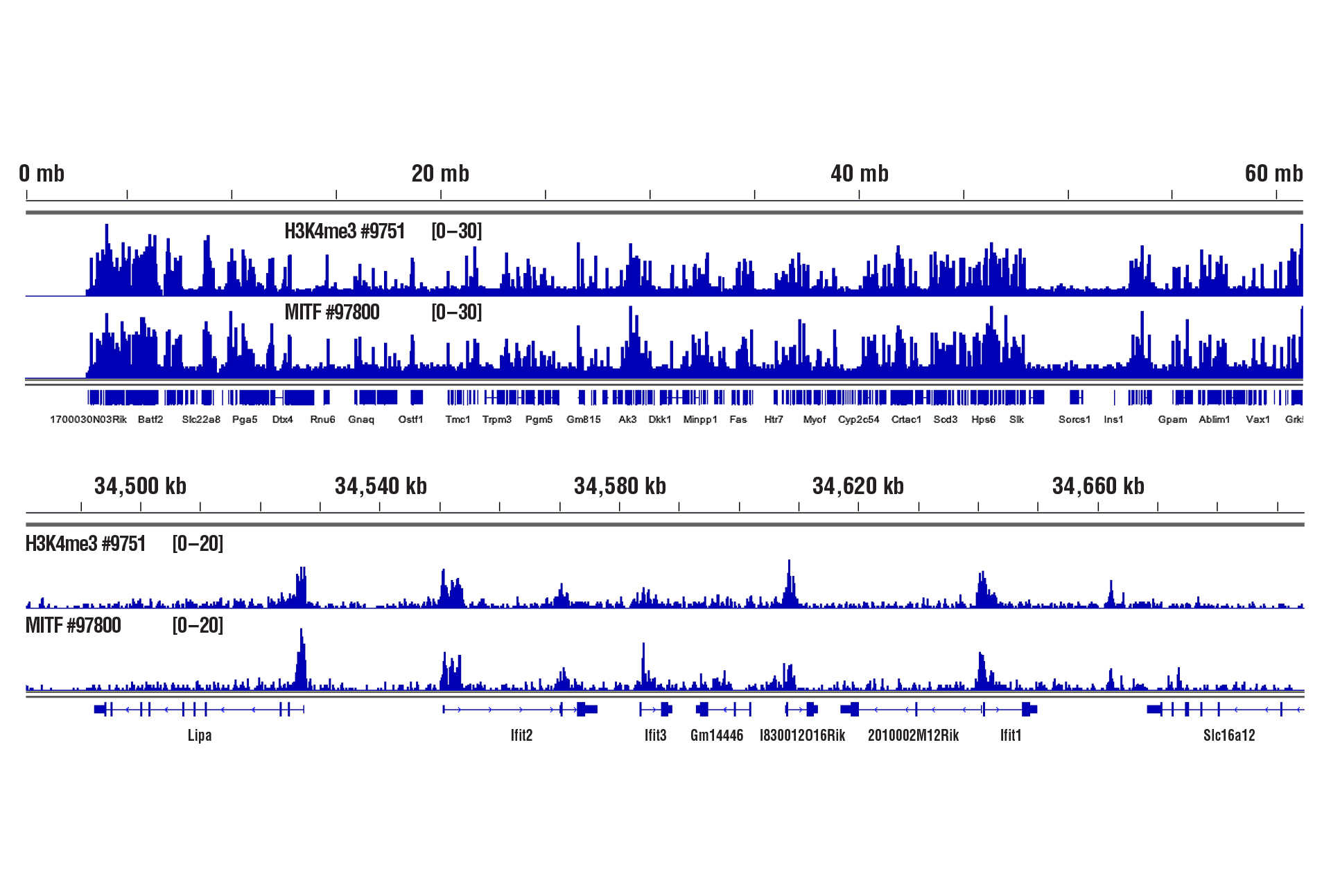
CUT&RUN was performed with 1 mg of lightly fixed mouse heart tissue (0.1% formaldehyde, 2 minutes) and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 or MITF (D3B4T) Rabbit mAb #97800, using CUT&RUN Assay Kit #86652. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. The figure shows binding of H3K4me3 and MITF across chromosome 19 (upper), including Ifit2 gene (lower).
Only 1 mg of brain tissue was required to analyze protein-DNA interactions for H3K4me3 and transcription factor Brn2/POU3F2 using our CUT&RUN Kit.
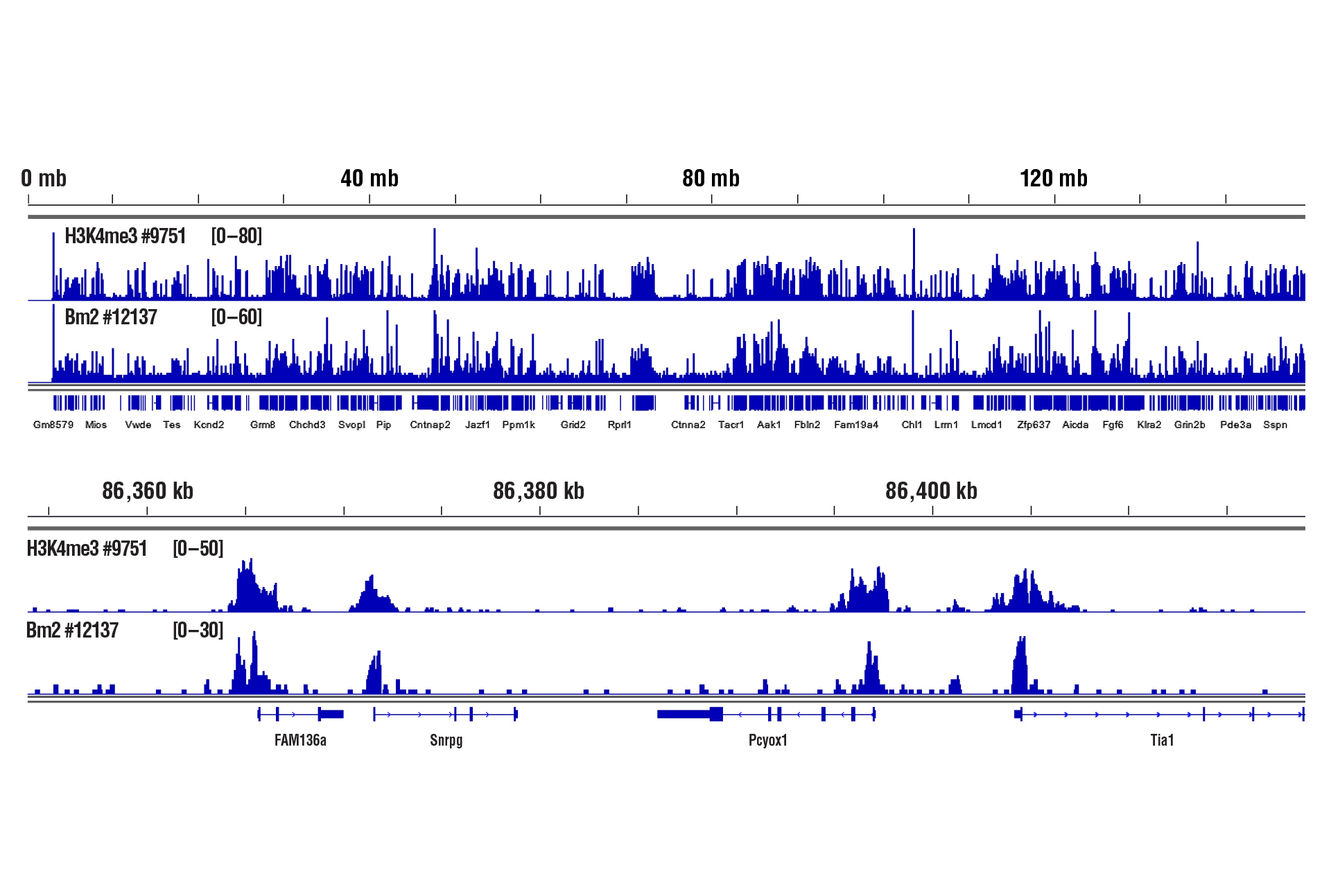
CUT&RUN was performed with 1 mg of lightly fixed mouse brain tissue (0.1% formaldehyde, 2 minutes) and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 or Brn2/POU3F2 (D2C1L) Rabbit mAb #12137, using CUT&RUN Assay Kit #86652. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. The figure shows binding of H3K4me3 and Brn2 across chromosome 6 (upper), including Snrpg gene (lower).
Only 1 mg of liver tissue was required to analyze protein-DNA interactions for H3K4me3 and the Glucocorticoid Receptor transcription factor using our CUT&RUN Kit.
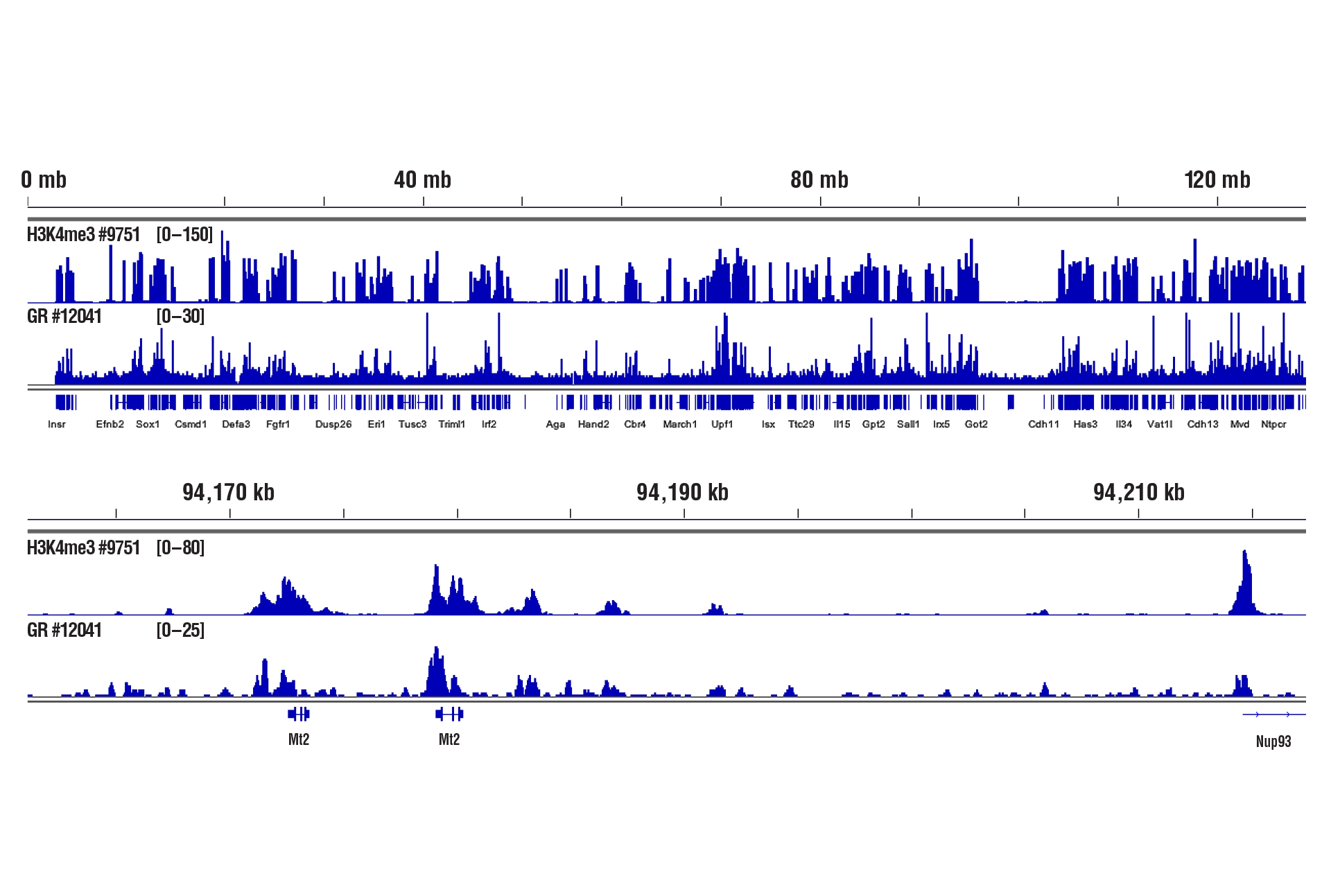
CUT&RUN was performed with only 1 mg of medium-fixed mouse liver tissue (0.1% formaldehyde, 10 minutes) and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 or Glucocorticoid Receptor (D6H2L) XP® Rabbit mAb #12041, using CUT&RUN Assay Kit CUT&RUN Assay Kit #86652. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. The figure shows binding of H3K4me3 and Glucocorticoid Receptor across chromosome 8 (upper), including Mt2 gene (lower).
Only 2.5 mg of liver tissue were required to analyze protein-DNA interactions for the cofactors SUZ12 and RING1B using our CUT&RUN Kit.
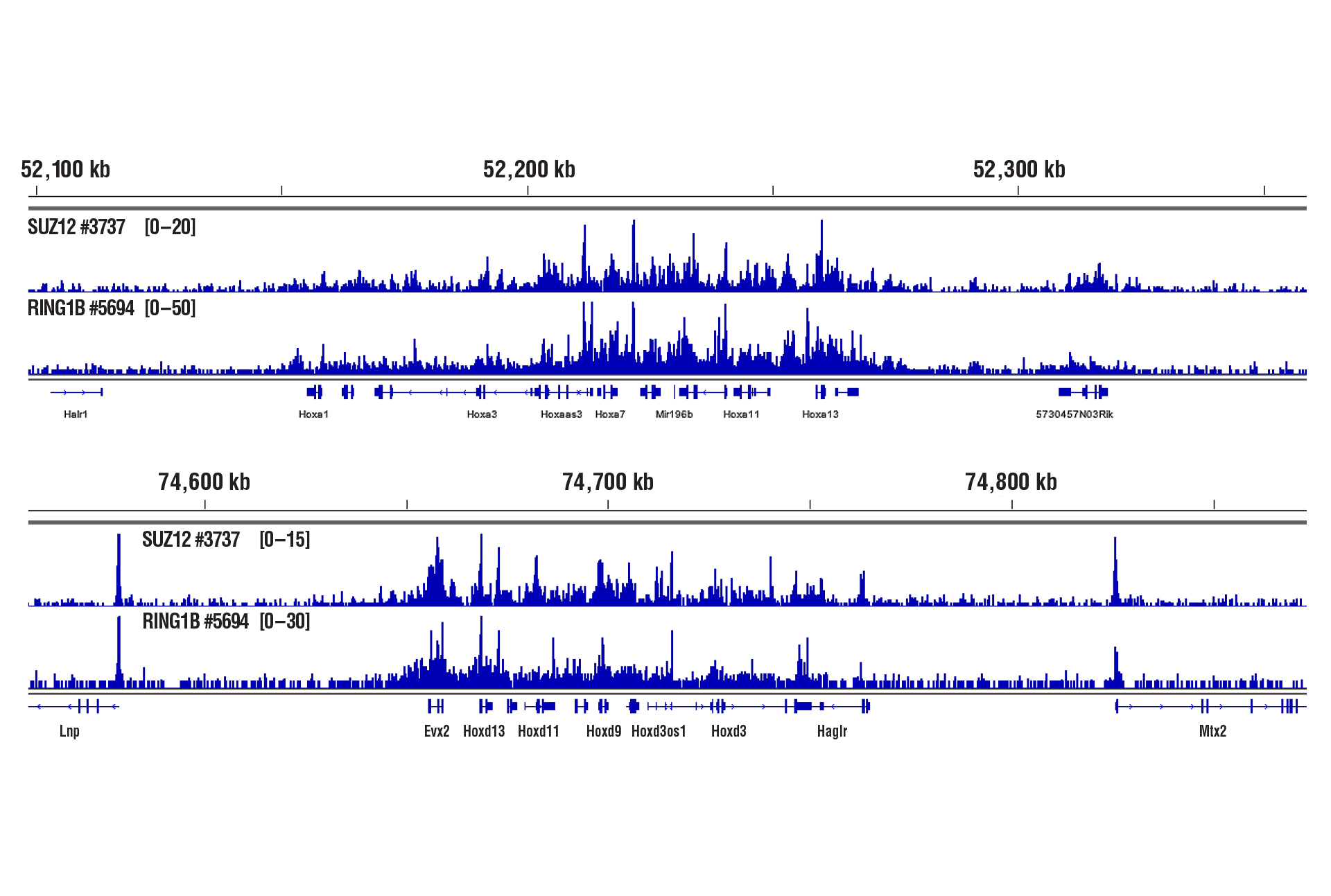
CUT&RUN was performed with 2.5 mg of medium-fixed mouse liver tissue (0.1% formaldehyde, 10 minutes) and SUZ12 (D39F6) XP® Rabbit mAb #3737 or RING1B (D22F2) XP® Rabbit mAb #5694, using CUT&RUN Assay Kit #86652. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. The figure shows binding of SUZ12 and RING1B across HoxA (upper) and HoxD (lower) gene regions.
Before CUT&RUN, analyzing protein-DNA interactions in primary cells, which aren’t immortalized, with ChIP was nearly impossible due to the challenges involved with trying to grow up enough cells.
The CUT&RUN Assay Kit #86652 significantly lowers the cell number requirement, making studying protein-DNA interactions in primary cells feasible. Don’t compromise your desired sample type just to meet cell number requirements.
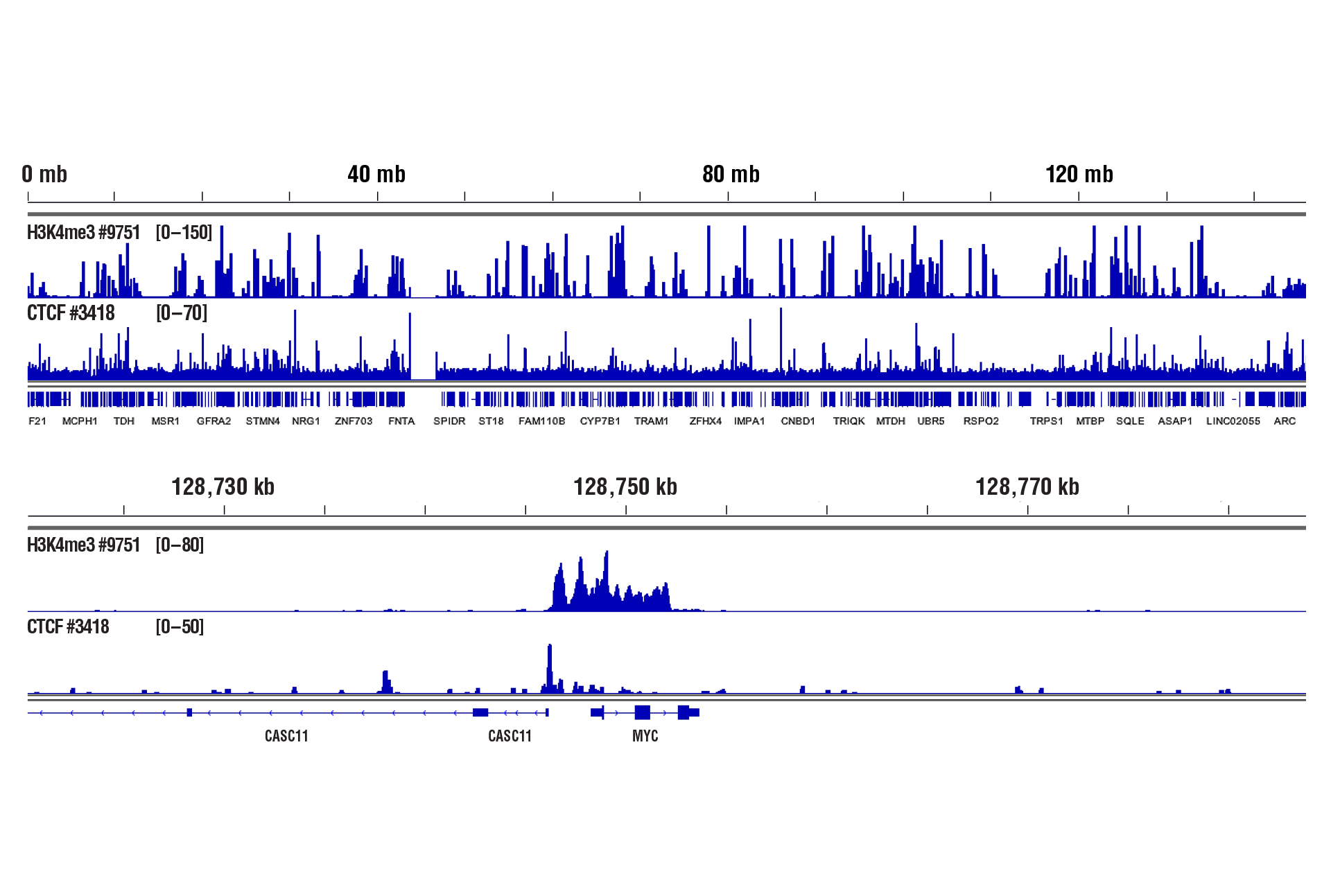
CUT&RUN was performed with 100,000 live human CD8+ T cells and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 or CTCF (D31H2) XP® Rabbit mAb #3418, using CUT&RUN Assay Kit #86652. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. The upper panel shows binding across chromosome 8, while the lower panel shows enrichment around the MYC gene, a known target of H3K4me3 and CTCF. See additional figure containing CUT&RUN-qPCR data in #3418 datasheet.
CST offers CUT&RUN reagents in a kit or a la carte, so you can order one kit to get all the necessary reagents or order just the reagents you need. They are all validated in-house to ensure you’ll get the high-quality reagents you’ve come to expect from CST.
| Catalog # | Product |
| 86652 | CUT&RUN Assay Kit |
| 40366 | CUT&RUN pAG-MNase and Spike-in DNA |
| 14209 | DNA Purification Buffers and Spin Columns (ChIP, CUT&RUN) |
| 88989 | SimpleChIP® Universal qPCR Master Mix |
| 56795 | DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795 |
| 47538 | Multiplex Oligos for Illumina (Dual Index Primers) (ChIP-seq, CUT&RUN) |
| 29580 | Multiplex Oligos for Illumina (Single Index Primers) (ChIP-seq, CUT&RUN) |
| 66362 | Rabbit (DA1E) mAb IgG XP® Isotype Control (CUT&RUN) |
| 93569 | Concanavalin A Magnetic Beads and Activation Buffer |
| 16359 | Digitonin Solution |
| 27287 | 100X Spermidine |
| 7012 | Protease Inhibitor Cocktail (200X) |
| 7013 | RNAse A (10 mg/ml) |
| 10012 | Proteinase K (20mg/ml) |
| 31415 | CUT&RUN 10X Wash Buffer |
| 15338 | CUT&RUN Antibody Binding Buffer |
| 48105 | CUT&RUN 4X Stop Buffer |
| 42015 | CUT&RUN DNA Extraction Buffer |