14 human cell lines commonly used in labs around the world were analyzed using TMT10plex™. A mixture of peptides from all 14 cell lines (numbered below) was used to standardize the quantitation across multiple 10-plexed experiments. Relative protein abundance in each cell line compared to the control mix was calculated and color-coded to reflect proteins that were higher abundance than control (green) or lower abundance compared to control (red).
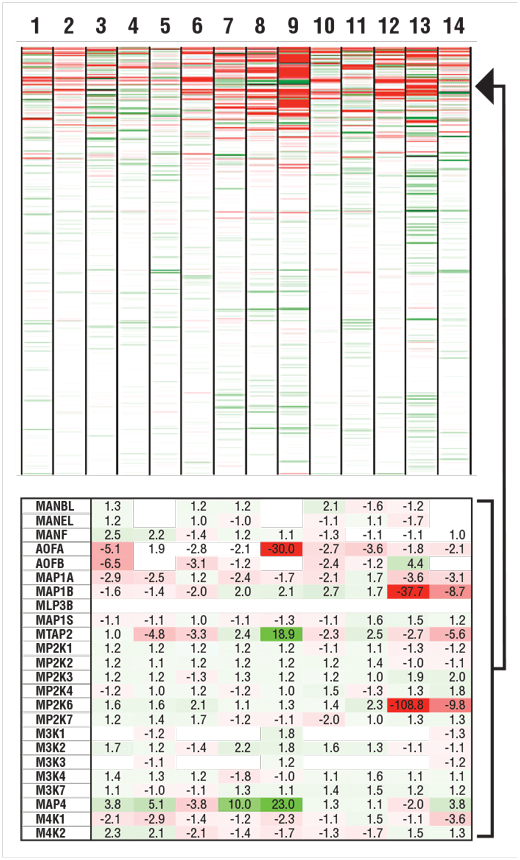
Heat map of color-coded relative protein abundances measured with TMT10plex™ across 14 human cell lines (numbered). Over 8000 proteins were quantified (upper), and a subset of data points shown with color coding and % change in abundance relative to control mix (lower).
Clients can view data globally using hierarchical clustering or other tools, and can review individual protein data in the tables.
TMT10plex™ data was compared to to western blots (WB) to validate protein abundance measurements. TMT™ quantitation shows good agreement with WB. FoxA2 and CD3ε expression levels varied with cell line, while Erk1 and Erk2 exhibited less cell line-dependent variation in expression.
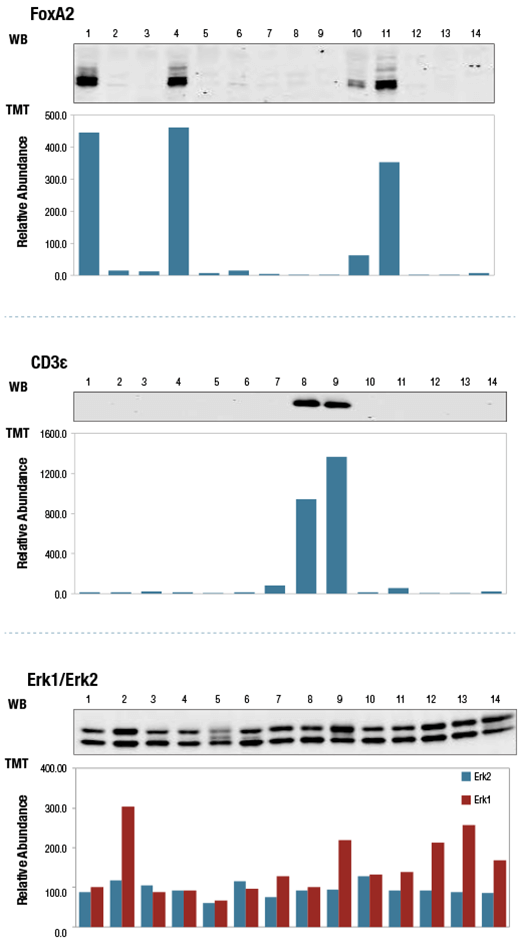
Relative protein abundance quantitation in 14 human cell lines (numbered) by TMT10plex™ profiling (bar graphs) and aligned western blots for FoxA2 (upper), CD3ε (middle) and Erk1/2 (lower).
These data demonstrate the ability of the TMT10plex™ Total Proteome method to provide detailed, quantitative information on relative protein abundance from multiplexed samples.
TMT10plex™ is a trademark of Thermo Fisher.