Mixed lineage leukemia is characterized by chromosomal translocations of the mixed lineage leukemia (MLL) gene to form fusion proteins with partner proteins. Many of these MLL-fusion partners belong to the super-elongation complex involved in regulating transcriptional elongation. MLL-fusions can create oncogenic transcription factors that cause aberrant transcriptional regulation of oncogenes or tumor suppressors to initiate oncogenesis.
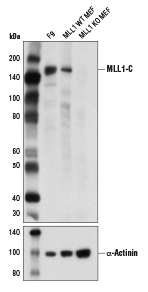
MLL1 is processed into two polypeptides, both which are subunits of the wild-type MLL complex. The amino terminus of MLL1 is what is included in the fusion proteins that cause leukemogenesis. We recommend using a carboxy-terminal specific antibody for detecting wild-type MLL1 and an amino-terminal specific antibody for detecting MLL-fusion proteins.
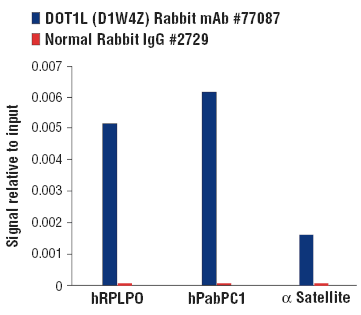
DOT1L interacts with MLL translocation partners in the super-elongation complex and is required for MLL-driven leukemiogenesis. DOT1L is thought to drive gene expression through the methylation of histone H3 at Lys79.
p300 and CBP are additional MLL-fusion proteins that can drive gene expression through histone acetylation.
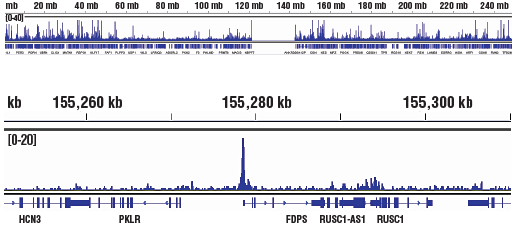
Changes in levels of these marks as detected by ChIP/ChIP-seq may indicate altered recruitment of MLL1, DOT1L, and CBP/p300 to target genes.